Quantasoft Software For Mac
Further, after a successful zagruki Mac OS X ustnavlivaem from a folder! Choose KernelBooter_kexts_title as in the picture, the rest of the default and copy the Extra folder from the USB device to the hard drive with. Mac OS X El Capitan 10.11.6 15G31 Intel USB 2017 – Freeware Sys │── Downloaded from Katcr.co.txt. Windows 10 and Apple's OS X El Capitan go head to head in nine rounds of combat. Here's why Microsoft's OS comes out on top. January 8th, 2017 at 2:53 pm. This feature is not available right now. Please try again later. OS X El Capitan remains available for Mac computers that can't upgrade to macOS Mojave, High Sierra, or Sierra, or that need to upgrade to El Capitan first. 
When Software Update says that your Mac is up to date, macOS and all of its apps are also up to date. That includes Safari, i Tunes, Books, Messages, Mail, Calendar, Photos, and FaceTime. To automatically install macOS updates in the future, including apps downloaded from the App Store, select ”Automatically keep my Mac up to date.”.
(425 bytes) The data referenced by this article are under copyright with the following copyright statement: Copyright: © 2016 Attali D et al. Data associated with the article are available under the terms of the Creative Commons Zero 'No rights reserved' data waiver (CC0 1.0 Public domain dedication). F1000Research: Dataset 1. Raw ddPCR data from application of the ddPCR assay against BRAF-V600 mutations, F1000Research: Dataset 2.
The set of exported CSV files of the data presented in., is also available as a sample dataset within the ddpcr package. To access the data via the web application, select the tab Use sample dataset, choose Large dataset, and then click Load data. To access the data in R, run the following command to store the dataset as a plate object: my_data.
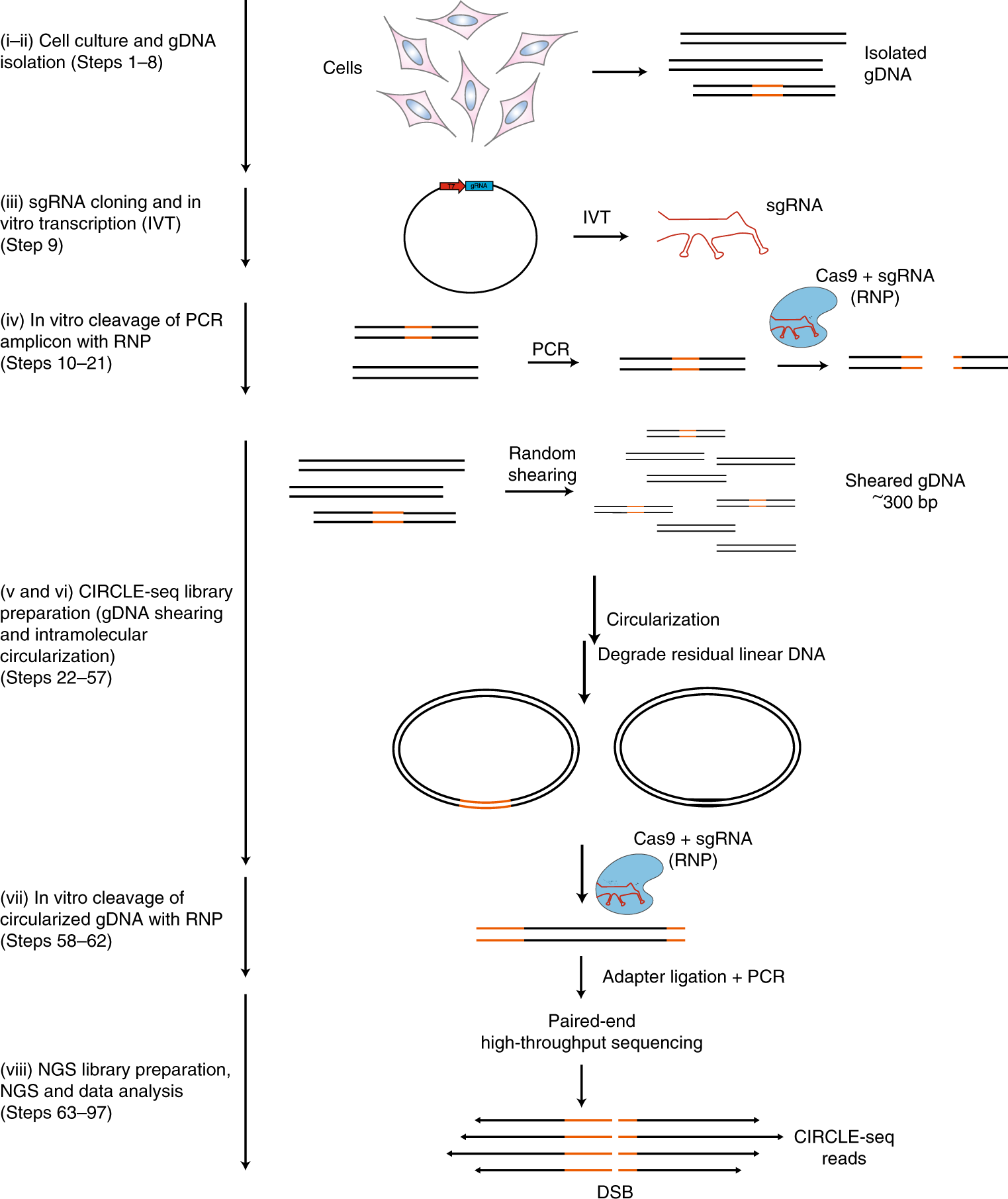
Droplet digital polymerase chain reaction (ddPCR) is a novel platform for exact quantification of DNA which holds great promise in clinical diagnostics. It is increasingly popular due to its digital nature, which provides more accurate quantification and higher sensitivity than traditional real-time PCR. However, clinical adoption has been slowed in part by the lack of software tools available for analyzing ddPCR data. Here, we present ddpcr – a new R package for ddPCR visualization and analysis. In addition, ddpcr includes a web application (powered by the Shiny R package) that allows users to analyze ddPCR data using an interactive graphical interface.
Introduction Droplet digital polymerase chain reaction (ddPCR) accurately quantifies targeted nucleic acid sequences (templates) by randomly partitioning sample DNA into isolated droplets, such that most droplets contain at most one template. The template within each droplet is then amplified and detected in a sequence-specific manner using a hydrolysis probe. The counting of droplets emitting a sequence-specific fluorescent signal permits the number of copies of that sequence present in the sample to be quantified with excellent sensitivity and precision. Different templates, such as wild-type and mutant alleles, may be quantified by using a uniquely labeled probe against each. The most commonly used reporter dyes on the probes are FAM (fluorescein) and HEX™, with the end-point fluorescence amplitudes for the two dyes measured by analyzing each droplet with a two-channel fluorescence detector. DdPCR data readily lends itself to visualization as a two-dimensional scatter plot ( ), in which the fluorescence amplitudes in both channels are plotted against each other for every droplet.
In a ddPCR experiment designed to quantify two different templates, droplets ideally segregate into unique groups (clusters) that may include HEX-positive, FAM-positive, double-positive, and double-negative (empty) clusters. For example, distinct FAM-positive, double-positive, and empty droplet clusters can be seen in. In practice, some droplets record an ambiguous set of fluorescent signals that fall between the distinct positive and negative populations. Such droplets are termed “rain” and can be observed between all clusters. By gating the droplets into groups based on their fluorescence signals, the exact number of template-positive droplets can be counted to provide exact quantification in a digital form. Comparison between droplet gating in ( A) QuantaSoft and ( B) ddpcr. Both tools analyzed the same ddPCR experiment (well F05) from an assay designed to quantify wild-type (double-positive) and mutant (FAM-positive) alleles of the BRAF gene.
( A) QuantaSoft failed to assign the double-positive and FAM-positive droplets into unique clusters, instead assigning all droplets recording a high FAM signal to a single cluster; ( B) ddpcr assigned droplets into one of three uniquely identified clusters (double-positive (green), FAM-positive (orange), and empty (black)), or rain (blue). Motivation Quantification of template abundance from raw ddPCR data begins with assigning each droplet to a unique cluster or to rain. The QuantaSoft program (Bio-Rad, Hercules, CA) is designed to perform these assignments either via manual gating, with the usual disadvantages of subjectivity and non-reproducibility, or automatic gating. The algorithm used in the latter case is proprietary and can produce unsatisfactory results, especially when applied to ddPCR data obtained from formalin-fixed paraffin-embedded (FFPE) samples, as exemplified in. Two third-party tools for automatic gating of ddPCR data have been described to date: ‘definetherain’ by Jones et al. And ’ddpcRquant’ by Trypsteen et al.